Luis' Shared Code
This page contains some of the programs that I've written over the last few years at the University of Michigan and I often use in my work. I'm happy to share my work with others, but I would like to ask that you send me an email if you download the programs, so that I know whether the stuff is useful. You can modify anything you want to suit your needs.
DISCLAIMER: I am not responsible for what happens to your data. Just because it works for me doesn't mean that it will work for you. Having said that, please let me know if you find bugs. Chances are you will find quirks here and there. Please send me an email if you find bugs and I'll see what I can do!
Luis' Tools
Both of the programs described here come bundled in a single tar file. Download the whole Toolbox.
Installation
This TAR file contains several tools for image display and analysis in matlab. There are a lot interdependencies between the functions, so I recommend that you download the whole toolbox to avoid problems. Some of these functions also use functions from SPM.
-
Download the .tar file (LuisTools.tar) and expand it in a directory of choice.
tar xvf LuisTools.tar
-
Add that directory to your Matlab path. For instructions on how to use a specific function from the toolbox, say, "blah.m", simply type (within matlab)
> help blah
and you will get instructions on the usage input and output.
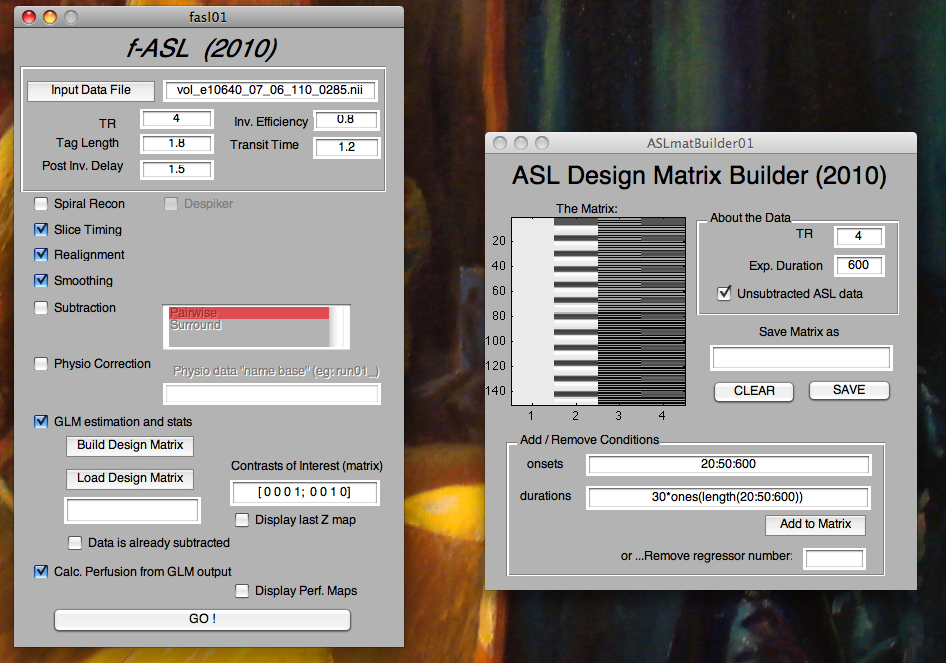
f-ASL
An interactive tool for analysis of arterial spin labeling functional MRI time series data. It includes preprocessing, physiological noise correction, OLS and GLS analysis, as well as quantification of perfusion.
Brief User Manual (f-ASL)
To start f-ASL, run Matlab and execute the command:
fasl01
A GUI interface will pop up. The general idea is that you provide a raw data file and check all the things that you want to do to the data. When you click on GO, the program will execute the options you checked thing at once from top to bottom. It’s really simple.
If you are going to do a GLM analysis, you will need to either load a design matrix from a text file. If you don't have one already made, then you can click the "Build Design Matrix" option to generate one - the ASL design matrix builder tool will pop up, and you can fill in the regressors one by one.
The nice features about this program are that
It has a tool for physiological noise correction using RETROICOR adapted for ASL data
is creates the design matrix for unsubtracted ASL data for you.
-
if analyzing unsubtracted data, the program will do GLS estimation (prewhitening of the data). See:
Mumford, J.A., Hernandez-Garcia, L., Lee, G.R., Nichols, T.E., 2006. Estimation efficiency and statistical power in arterial spin labeling fMRI. Neuroimage 33, 103-114.
-
After GLM estimation, it converts parameter estimates into meaningful perfusion effects in units of perfusion (ml/min/100g). Please see the forthcoming paper (you can already see in in PubMed though):
Hernandez-Garcia L, Jahanian H, Rowe DB: Quantitative Analysis of Arterial Spin Labeling FMRI Data Using a General Linear Model. Magnetic Resonance Imaging, 28 (7), Pages 919-927, 2010. [PMID: 20456889]
-
If you know how to program matlab, you can run it in a batch mode (skipping the GUI) by calling the "master program" from a script. FASL is just a nice GUI wrapper around this other program. The program file is asl_spm01.m. To use it, all you have to do is update the contents of the structure "args" and call
asl_spm01(args)
Type help asl_spm01 to get a complete listing of the contents of "args" and how to use it.
Important Notes
Some things are hard coded, and you may need to get under the hood, or contact me about how to change them. IN particular, the physio correction process is tailored to our scanner's sampling rate and our naming scheme for the physiological data files. Right now, you need to get into the code and do some editing to change that. (Not being familiar with too many scanners, I’m not sure how to do a generic one, so in the mean time you can email me and I'll help).
The reconstruction code is also set up to reconstruct images from our scanner and our specific pulse sequence, so you'll want to skip that part and start out with already reconstructed images from your site. If you're a regular matlab hacker, it should be easy to stick in your own reconstruction code into it.
The slice timing correction interpolates the data to the last slice in the acquisition. Make sure you know your know your acquisition timing parameters.
The smoothing tool is set up to use a Gaussian kernel with 3 voxels width.
The realignment is done by SPM 5 and the data get resliced.
The quantifiation model assumes you are running a continuous type of ASL scheme, like pseudo CASL.
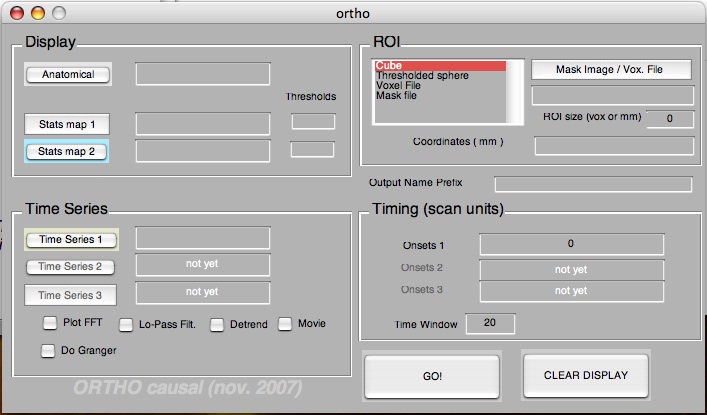
Ortho
An interactive tool for visualization and ROI analysis of FMRI time series data. This tool also includes Granger Causality Analysis (use at your own risk).
Brief User Manual (Ortho)
Starting Ortho
Run Matlab and execute the command:
ortho
(The GUI portion does not work with prior versions to Matlab 7, but you can still run ortho as a command line tool, though!)
If you want to run ortho without the GUI, take a look at the file ortho_nogui.m . You can edit that file by filling in the fields that you need, and use it as a "fake GUI". Once you have filled it in, all you have to do is execute it.
Similarly, If you want to use the programs in batch mode to run through all your subjects, edit the file ortho_batch.m and execute it.
But, if you are one of the lucky ones to have Matlab 7 and later and you are using the GUI, the interface looks like the picture on the right.
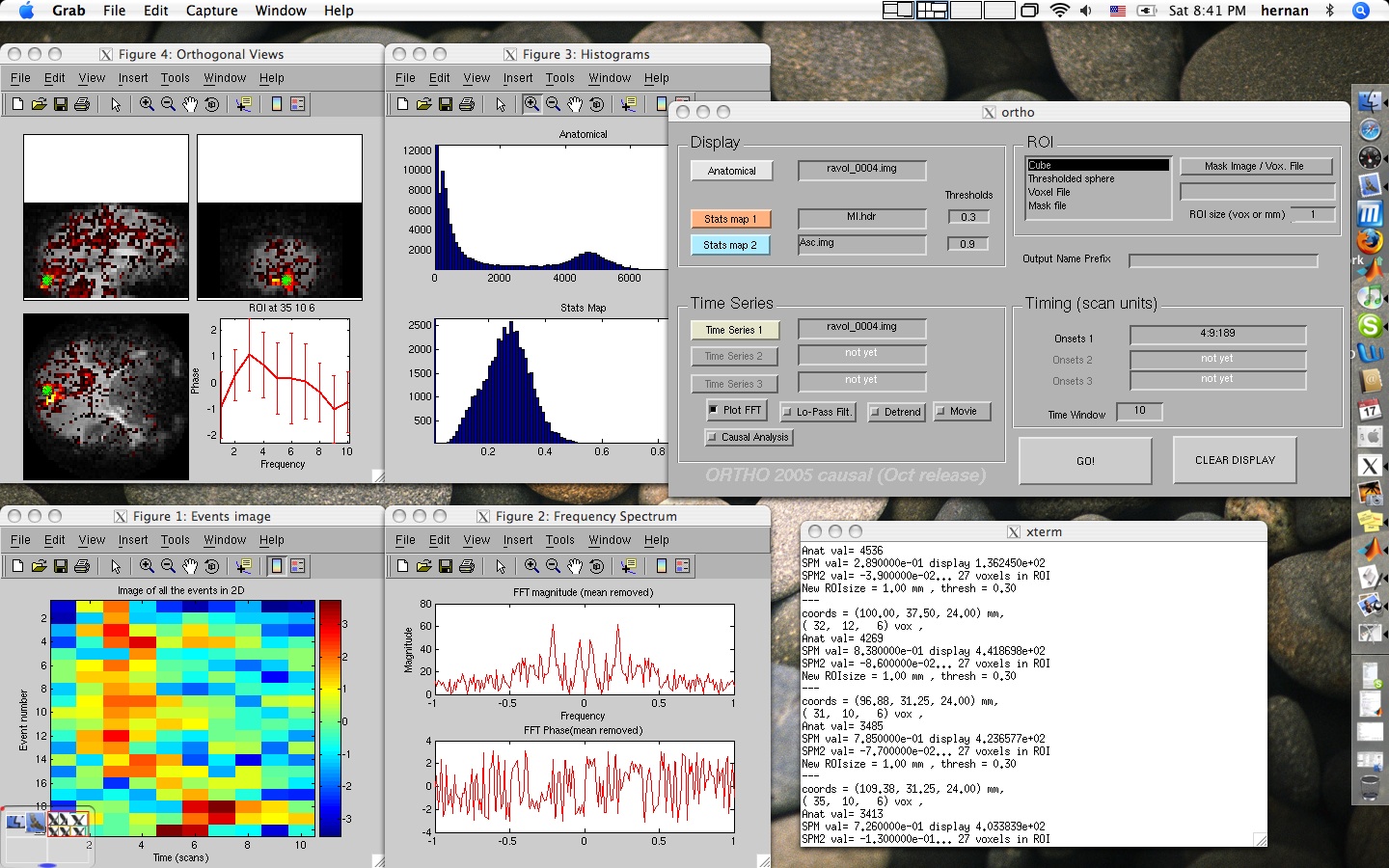
Using Ortho
Ortho works on Analyze format files (.img) and NIFTI (.nii. nii.gz). Please note that ORTHO disregards the .mat files that are generated by SPM. It does use the header information like origins, voxel sizes and Scaling Factor.
Now that ORTHO is running, its main functions are to
- display the images in orthogonal views,
- overlay statistical maps on top of them and
- extract time series from selected voxels. You can easily select the type or ROI you want.
- show you pixel values of the anatomical and the statistical images.
ORTHO can also (if you check the options)
- average events from a time series,
- show you a movie of the image in orthogonal sections.
- filter the data,
- display the temporal frequency content. The user is allowed to navigate the images interactively, and ORTHO updates the time series display accordingly.
- save the time series data into ASCII files.
- ortho is VERY easy to run as a batch script. Type " help ortho2005 " at the matlab command prompt for details.
Ortho's interface is designed so that when you press the GO button, it displays as much as it can with whatever fields are filled in by the user, (... but some combinations are not implemented yet). ORTHO shows you the images and it updates the display interactively as you select voxels by clicking on them. Information about the pixel (or pixels) gets displayed in the matlab command window. When you RIGHT CLICK, all that information gets put into ASCII files.
Here is a brief description of how to do a few basic things using the GUI:
-
Extracting and Looking at a time series:
If you fill in the "time series" only, you will get a plot of the time series at whatever voxel you click on. To fill in the "Time Series" field, select one of the images in the time series. ORTHO will assume that they all are named the same except for the numbers at the end of the name. eg-
vol_e2343_10_12_03_0001.img
vol_e2343_10_12_03_0002.img
...etcIf you have a single 4D file, ORTHO will recognize it and read it in as well. If you fill in the "anatomical" image only, you will get a display of the image you select along with a nice histogram of the pixels values.
-
Stuff you can do to the time series:
You can also add detrending (3rd order polynomial) and gaussian filtering to the time series by checking the appropriate box. The filter parameters are hard-coded. They should be user defined, but that's not implemented yet... sorry... it's easy to hack if you have the inclination, though...
If you click with the RIGHT mouse button, the extracted waveforms get stored as ASCII files for easy access outside of matlab.
You can display the FFT's magnitude and phase in a separate window by checking the "FFT" box (near the bottom)
If you provide a set of onset times, and a time window, you will get trial averages over the desired widow of time. Note - ortho looks at time in units of scans - does not know your sampling rate or TR. It also displays a nice image of a matrix made up of the individual events that you are averaging. This is really handy because you can notice changes in the events from trial to trial, things like habituation or drifts in HRF parameters, errors in the timing of the experiment...
You can also look at a movie of the time series through the selected location be checking the "Movie" checkbox
-
Ovelaying a Statistical Map:
If you fill in the "anatomical" and "stats map 1", you will get a display of the anatomical image with the statistical map overlayed on it. You can select the threshold by filliing in the threshold value field. These images must be coregistered previously. They can have different resolutions, but they need to be in the same orientation with the same origin. Remember that ORTHO does not use the .mat files generated by SPM's coregistration so it doesn't apply the transformations on the fly the way SPM does.
You can also display two statistical maps to find overlapping voxels by filling in "stats map 2" as well. Overlapping pixels in the statistical maps are shown in green.
You can change the threshold interactively by filling in a new value in the interface and clicking on a pixel to update the display.
-
Selecting ROI types:
If you fill in the "Anatomical", "Stats Maps" and "Time Series", you can also select a specfic set of voxels to extract the information from. The values in those voxels get averaged together. You can choose what kind of ROI to use in the upper right side of the interface:
Cube: means that you get the voxel you click on plus the number of neighbors in each direction specified in the field labeled "ROI size". The default is cube with zero neighbors.
Thresholded Sphere: means a sphere with the radius in mm specified in the field labeled ROI size.
Mask File: means an Analyze format image in which the voxels of interest have value 1. All others have value of zero.
Voxel File means that the ROI consists of a list of voxels specified in an ASCII file, which you have to select by clicking the "Mask File / Vox. File" Button, or just typing in the file name in the field below that button. About the aforementioned voxel file, it needs to be an ascii (text) file that looks like this example:
12 43 45
12 44 46
23 45 89
...etc.Those coordinates are in voxel units and they are relative to the first voxel in the file - the one in the lower left corner, NOT the orgin!
-
Computing Granger Causality Maps:
There is a CheckBox at the bottom of the GUI that says "Do Granger". IF this is checked, as soon as you RIGHT-click on a pixel, the program will compute the Granger Causality F score between the current ROI and the rest of the image. The resulting files are named:
grangerFab - F score of the influence of seed on each voxel
grangerFba - F score of the influence of each voxel on the seed.
grangerF - difference between the two above: a measure of the directionality of the relationship. If it's positive, it means that the seed granger-Causes the voxel in question.
grangerlog10P - a p-value associated with the F score above (I'm not very confident on this p-value computation though)You also get histograms of the different outputs.
Be careful with Granger Causality; Whether it’s appropriate for BOLD time series data is not clear at all!